Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About me
This is a page not in th emain menu
Published:
This post is aimed at those who want to understand the mathematical framework of denoising diffusion probabilistic model and its implementation in deep learning.
Published:
In this post I explain the mathematics behind conditional variational autoencoders and the differences with conventional variational autoencoders.
Published:
This post is aimed at those who want to understand the mathematical framework of variational autoencoders and its implementation in deep learning.
Published:
The aim of this post is to present the key stages/concepts of contrastive learning.
Published:
Are you interested in understanding the mathematics that underlie the transformer? If so, this post is tailored for you!
Published in Plos one, 2022
This paper introduces CMRSegTools: an open-source application software designed for the segmentation and quantification of myocardial infarct lesion enabling full access to state-of-the-art segmentation methods and parameters, easy integration of new algorithms and standardised results sharing.
Recommended citation: Romero, William (2022). "CMRSegTools: An open-source software enabling reproducible research in segmentation of acute myocardial infarct in CMR images." Plos one. 17(9). https://hal.science/hal-04215855
Published in IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 2022
We present a numerical framework for generating clinical-like Color Doppler imaging. Synthetic blood vector fields were obtained from a patient-specific computational fluid dynamics CFD model.
Recommended citation: Sun, Yunyun (2022). "A Pipeline for the Generation of Synthetic Cardiac Color Doppler." IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. 69(3). https://cnrs.hal.science/hal-03538666
Published in medRxiv, 2022
We evaluate the accuracy and reproducibility of 2D echocardiography left ventricular volumes and ejection fraction estimates by Deep Learning versus manual contouring and against CMR.
Recommended citation: Saloux, Eric (2022). "Deep Learning vs manual techniques for assessing left ventricular ejection fraction in 2D echocardiography: validation against CMR." medRxiv. https://www.medrxiv.org/content/10.1101/2022.07.26.22278059v1
Published in IEEE Transactions on Medical Imaging, 2022
We propose a novel deep learning solution for motion estimation in echocardiography. In parallel, we designed a novel simulation pipeline allowing the generation of a large amount of realistic B-mode sequences.
Recommended citation: Evain, Ewan (2022). "Motion Estimation by Deep Learning in 2D Echocardiography: Synthetic Dataset and Validation." IEEE Transactions on Medical Imaging. 41(8). https://hal.science/hal-03603014
Published in Medical Image Computing and Computer Assisted Intervention -- MICCAI, 2022
We propose a method called CRISP for uncertainty prediction of image segmentation. CRISP implements a contrastive method to learn a joint latent space which encodes a distribution of valid segmentations and their corresponding images.
Recommended citation: Judge, Thierry (2022). "CRISP - Reliable Uncertainty Estimation for Medical Image Segmentation." Medical Image Computing and Computer Assisted Intervention -- MICCAI. https://hal.science/hal-04215854
Published in IEEE Transactions on Medical Imaging, 2022
We propose a framework to learn the 2D+time apical long-axis cardiac shape such that the segmented sequences can benefit from temporal and anatomical consistency constraints.
Recommended citation: Painchaud, Nathan (2022). "Echocardiography Segmentation With Enforced Temporal Consistency." IEEE Transactions on Medical Imaging. 41(10). https://hal.science/hal-03672999
Published in IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 2023
We developed an unfolded primal-dual network to unwrap (dealias) color Doppler echocardiographic images and compared its effectiveness against two state-of-the-art segmentation approaches based on nnU-Net and transformer models.
Recommended citation: Ling, Hang Jung (2023). "Phase unwrapping of color Doppler echocardiography using deep learning." IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. 70(8). https://hal.science/hal-04142824v2
Published in IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 2023
We developped a complex-weighted convolutional neural network (CNN) for ultrasound images. We showed that our method allows reconstructing high-quality static images while maintaining the capability of tracking cardiac motion
Recommended citation: Lu, Jingfeng (2023). "Ultrafast Cardiac Imaging Using Deep Learning For Speckle-Tracking Echocardiography." IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. https://arxiv.org/abs/2306.14265
Published in Medical Image Computing and Computer Assisted Intervention -- MICCAI, 2023
We propose explicitly modeling location uncertainty by redefining the segmentation task as contour regression, providing improved performance and interpretability.
Recommended citation: Judge, Thierry (2023). "Asymmetric Contour Uncertainty Estimation for Medical Image Segmentation." Medical Image Computing and Computer Assisted Intervention -- MICCAI. https://hal.science/hal-04243975v1
Published in IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 2024
We propose novel alternatives to the traditional intraventricular vector flow mapping (iVFM) optimization scheme by utilizing physics-informed neural networks (PINNs) and a physics-guided nnU-Net-based supervised approach.
Recommended citation: Ling, Hang Jung (2024). "Physics-Guided Neural Networks for Intraventricular Vector Flow Mapping." IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. https://arxiv.org/abs/2403.13040
Published in Medical Image Computing and Computer Assisted Intervention -- MICCAI, 2024
We present an innovative reinforcement learning framework for optimal domain adaptation in medical imaging segmentation. This framework reduces the need to otherwise incorporate large expertly annotated datasets in the target domain, and eliminates the need for lengthy manual human review.
Recommended citation: Judge, Arnaud (2024). "Domain Adaptation of Echocardiography Segmentation Via Reinforcement Learning." Medical Image Computing and Computer Assisted Intervention -- MICCAI. https://arxiv.org/pdf/2406.17902
Published in Medical Imaging with Deep Learning -- MIDL, 2025
We present a fusion strategy based on an asymmetric scheme designed to integrate patient data with information extracted from images. The model is validated on a dataset of 239 patients for characterizing hypertension severity.
Recommended citation: Stym-Popper, Jeremie (2025). "DAFTED: Decoupled Asymmetric Fusion of Tabular and Echocardiographic Data for Cardiac Hypertension Diagnosis." Medical Imaging with Deep Learning -- MIDL. https://openreview.net/pdf?id=ghhGImwv07
Published in Medical Image Computing and Computer Assisted Intervention -- MICCAI, 2025
We develop a methodology to evaluate segmentation methods based on the generation of realistic synthetic images through a diffusion network with explicitly controlled biomarker values.
Recommended citation: Deleat-besson, Romain (2025). "Controllable latent diffusion model to evaluate the performance of cardiac segmentation methods." Medical Image Computing and Computer Assisted Intervention -- MICCAI.
Published in Statistical Atlases and Computational Modeling of the Heart workshop -- STACOM, 2025
We evaluate MedSAM and SamMed2D for segmenting the left ventricle and myocardium in LGE MR images from a private dataset comprising 135 patients.
Recommended citation: Goujat, Celia (2025). "Medical SAM for LGE-MRI cardiac segmentation: promise or hype?" Statistical Atlases and Computational Modeling of the Heart workshop -- STACOM. https://hal.science/CREATIS/hal-05242396v1
Published in IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 2025
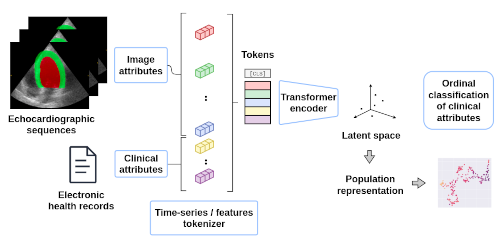
We propose a method that considers all descriptors extracted from medical records and echocardiograms to learn the representation of a cardiovascular pathology with a difficult-to-characterize continuum, namely hypertension.
Recommended citation: Painchaud, Nathan (2025). "." IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. https://arxiv.org/abs/2401.07796
Published in IEEE Journal of Biomedical and Health Informatics, 2025
We propose two deep learning methods to estimate global and segmental longitudinal strain of the left ventricle in transesophageal echocardiography. These methods are trained exclusively from realistic simulations and showed promising clinical applicability.
Recommended citation: Austlid Taskén, Anders (2025). "." IEEE Journal of Biomedical and Health Informatics. https://arxiv.org/abs/2511.02210
We developed a transformer-based approach to efficiently merge information extracted from echocardiographic image sequences and data from electronic health records to learn the continuous representation of patients with hypertension.
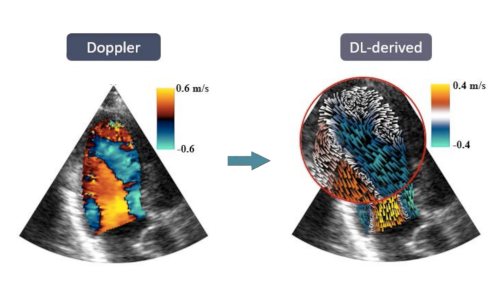
We developed a physics-informed deep learning method to estimate vector flow mapping from color Doppler imaging. This strategy has the potential to facilitate the analysis of the blood flow during clinical exams.
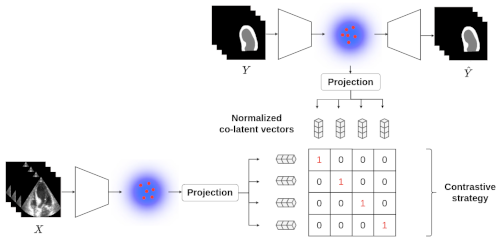
We have developed a generic method for estimating aleatoric uncertainty based on contrastive modeling.
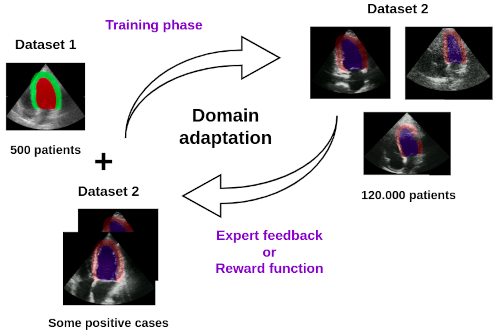
We are exploring domain adaptation strategies using reinforcement learning to enhance the generalizability of our segmentation models on echocardiographic images
We have developed a simulation pipeline to generate realistic synthetic echocardiographic sequences. This pipeline is currently being used to generate a large-scale cohort of virtual patients to feed deep learning methods.
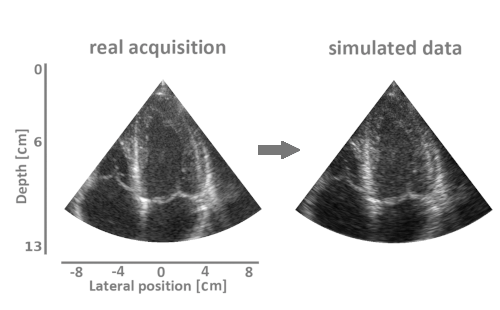
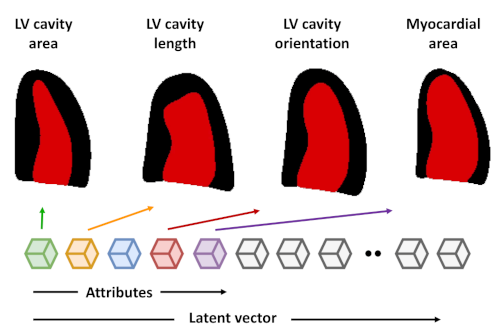
We have developed a strategy based on learning the representation of plausible cardiac shapes to efficiently generate gold standard annotations. This pipeline has been successfully applied to two other tasks: the simulation of realistic echocardiographic sequences and the generalization of segmentation tools.
Published:
In this presentation, I taught the formalism of the variational auto-encoder and emphasized the importance of creating specialized latent spaces using various examples. I then described the auto-encoder formalism and highlighted the additional advantages of the variational approach. I concluded by showing how we have exploited this formalism in order to i) provide guarantees on the segmented anatomical structures; ii) provide guarantees on the temporal consistency of the segmented shapes; iii) model the uncertainty of the segmentation algorithm.
Published:
Deep learning methods have allowed major advances in many specific areas such as echocardiogram analysis, view classification, recognition and segmentation of anatomical structures, user guidance or even automatic report generation. Moreover, the combination of these different techniques allows the deployment of complete and fully automated processing chains, making it possible to increase the reliability of clinical measurements and facilitate the use of ultrasound scanners outside of hospitals. In this talk, I described the various advances we have made using the deep learning paradigm for echocardigraphy image segmentation.
Published:
In recent years, artificial intelligence (AI) techniques have shown remarkable success in the field of medical imaging. While supervised learning approaches currently stand as the most effective methods, they rely heavily on ground truth data usually obtained from manual annotations, which often poses challenges across diverse applications. In this presentation, I introduced a comprehensive pipeline aimed at generating realistic simulations of echocardiographic image sequences to be used as inputs for supervised learning algorithms. I showcased their application in two different areas: tissue motion estimation in traditional imaging and ultra-fast cardiac image reconstruction.
Published:
AI paradigm now enables automatic and robust extraction of cardiac function descriptors from echocardiographic sequences, such as ejection fraction or strain. These descriptors provide fine-grained information that physicians consider, in conjunction with more global variables from the clinical record, to assess patients’ condition. In this presentation, I introduced a recently developed transformer-based framework that takes into account all descriptors extracted from medical records and echocardiograms. The framework aims to learn the representation of a challenging cardiovascular pathology, specifically hypertension. I showed that for descriptors whose interactions with hypertension are well documented, patterns are consistent with prior physiological knowledge, paving the way for further clinical studies to better understand this disease.
Published:
AI tools have demonstrated their efficiency in numerous cardiac applications for several years now. In this presentation, I describe (in french) four examples of applications where AI has revolutionized the status quo and enabled significant advancements towards precise, robust, and reproducible automation of routine clinical measurements, i.e. standardized acquisitions, extraction of anatomical indices, extraction of functional indices and uncertainty modeling
Published:
For several years now, deep learning (DL) techniques have been successfully applied to the segmentation of medical imaging. Several pilot studies initially showed the superiority of DL over conventional methods using databases of around one hundred patients. However, these initial results have raised other issues. Firstly, the generalization of DL methods to larger databases with high variability in shapes, vendors, image quality, and pathologies poses a challenge. Secondly, there is difficulty in producing reliable expert annotations on large databases. These challenges cast doubt on the ability of DL methods to provide a definitive solution to the segmentation problem in medical imaging. However, recent advancements in artificial intelligence (AI) research have revolutionized computer vision and image processing. This has led to the development of new powerful and more generic tools such as foundation models and reinforcement learning methods. Will the application of these tools in our field lead to the definitive resolution of segmentation in medical imaging in the near future? This is the burning question that I will address in my presentation.
Published:
Diffusion-based generative models have recently emerged as a powerful framework for modeling complex data distributions, especially in high-dimensional domains like medical imaging. In this lecture, I introduced the core concepts behind Denoising Diffusion Probabilistic Models (DDPM), a prominent class of diffusion models grounded in stochastic processes. I then illustrated their practical relevance through applications to data augmentation, semantic segmentation, and anomaly detection, highlighting how such models can generate anatomically plausible and diverse samples.
Published:
Generative AI has become a key paradigm in recent years, especially in the fields of natural language processing (NLP) and imaging, with the emergence of flagship models such as ChatGPT, DeepSeek, DALL-E, and MidJourney. However, its potential goes far beyond consumer applications: in medical imaging, generative AI plays a crucial role, particularly for data augmentation, anomaly detecetion, segmentation or image denoising. In this course, I explore the main generative AI approaches applied to medical image synthesis, with a specific focus on VAEs (Variational Autoencoders), and diffusion models. The goal is to provide a thorough understanding of these techniques and their relevance in the biomedical field. The course include interactive notebooks to illustrate these concepts in practice and allow hands-on experimentation with various approaches.
Course, INSA, university of Lyon, electrical department, 2023
French course on signal modeling. The following aspects are covered: basic signals / elementary operations, linear time-invariant systems, Fourier transform.
[pdf-fr]
Course, INSA, university of Lyon, electrical department, 2023
French course on signal sampling and quantization. The following aspects are covered: modeling of the sampling and quantization operations, energy assessment of the acquisition chain.
[pdf-fr]
Course, INSA, university of Lyon, electrical department, 2023
French course on the characterization of random signals. The following aspects are covered: deterministic vs. random signals, cumulative distribution function, probability density function, statistical moments, autocorrelation function, and power spectral density.
[pdf-fr] [code 1]
Course, INSA, university of Lyon, electrical department, 2023
French course on neural network. The following aspects are covered: modeling of a perceptron, modeling of a neural network, activation functions, backpropagation principle, loss function, gradient descent needs.
[pdf-fr] [code]
Course, INSA, university of Lyon, electrical department, 2023
French course on the optimization steps involved in neural networks. The following aspects are covered: mini-batch gradient descent, momentum/RMSprop/ADAM scheme, batch normalization, dropout, training/validation/test scheme.
[pdf-fr]
Course, INSA, university of Lyon, electrical department, 2023
French course on convolutional neural network. The following aspects are covered: convolutional layer, feature maps, receptive field, classification, segmentation.
[pdf-fr]
Course, INSA, university of Lyon, electrical department, 2024
Course on variational auto-encoder networks. The following aspects are covered: fundamental concepts, Kullback-Liebler divergence, variational inference, VAE architectures.
[pdf-fr] [pdf-en] [code 1] [code 2] [code 3] [code 4] [post]
Course, INSA, university of Lyon, electrical department, 2024
French course on transformers. The following aspects are covered: tokenisation, positional embeding, encoding blocs, self-attention module, multi-head attention, vision transformer.
[pdf-fr] [pdf-en] [code 1] [post]